Working with HLS datasets: cloud cover, GeoTIFFs, and the QA band¶
This guide shows how to use some of the utilities implemented in nasa_hls for working with NASA’s Harmonized Landsat and Sentinel-2 Project (https://hls.gsfc.nasa.gov/) data. It covers the follwoing topics:
Derive the cloud cover percentage from the metadata of a HDF file.
Convert HDF datasets to single layer GeoTIFFs.
Convert the binary QA layer in a clear-sky mask.
[1]:
%load_ext autoreload
%autoreload 2
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from pathlib import Path
import pandas as pd
import rasterio
import nasa_hls
The sample data¶
We assume the data that has been created wiht the ‘Query and download HLS datasets with nasa_hls’ guide.
[2]:
hdf_dir = Path("./xxx_uncontrolled_hls/downloads")
geotiff_dir = Path("./xxx_uncontrolled_hls/geotiffs")
import pandas as pd
df_downloaded = pd.read_csv("./xxx_uncontrolled_hls/downloads/df_downloads.csv", index_col="id")
display(df_downloaded)
| product | tile | date | url | year | month | day | path | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| HLS.L30.T32UNU.2018092.v1.4 | L30 | 32UNU | 2018-04-02 | https://hls.gsfc.nasa.gov/data/v1.4/L30/2018/3... | 2018 | 4 | 2 | ./xxx_uncontrolled_hls/downloads/HLS.L30.T32UN... |
| HLS.S30.T32UNU.2018092.v1.4 | S30 | 32UNU | 2018-04-02 | https://hls.gsfc.nasa.gov/data/v1.4/S30/2018/3... | 2018 | 4 | 2 | ./xxx_uncontrolled_hls/downloads/HLS.S30.T32UN... |
| HLS.L30.T32UPU.2018092.v1.4 | L30 | 32UPU | 2018-04-02 | https://hls.gsfc.nasa.gov/data/v1.4/L30/2018/3... | 2018 | 4 | 2 | ./xxx_uncontrolled_hls/downloads/HLS.L30.T32UP... |
| HLS.S30.T32UPU.2018092.v1.4 | S30 | 32UPU | 2018-04-02 | https://hls.gsfc.nasa.gov/data/v1.4/S30/2018/3... | 2018 | 4 | 2 | ./xxx_uncontrolled_hls/downloads/HLS.S30.T32UP... |
| HLS.L30.T32UPU.2018099.v1.4 | L30 | 32UPU | 2018-04-09 | https://hls.gsfc.nasa.gov/data/v1.4/L30/2018/3... | 2018 | 4 | 9 | ./xxx_uncontrolled_hls/downloads/HLS.L30.T32UP... |
| HLS.S30.T32UPU.2018099.v1.4 | S30 | 32UPU | 2018-04-09 | https://hls.gsfc.nasa.gov/data/v1.4/S30/2018/3... | 2018 | 4 | 9 | ./xxx_uncontrolled_hls/downloads/HLS.S30.T32UP... |
| HLS.L30.T32UNU.2018115.v1.4 | L30 | 32UNU | 2018-04-25 | https://hls.gsfc.nasa.gov/data/v1.4/L30/2018/3... | 2018 | 4 | 25 | ./xxx_uncontrolled_hls/downloads/HLS.L30.T32UN... |
| HLS.S30.T32UNU.2018115.v1.4 | S30 | 32UNU | 2018-04-25 | https://hls.gsfc.nasa.gov/data/v1.4/S30/2018/3... | 2018 | 4 | 25 | ./xxx_uncontrolled_hls/downloads/HLS.S30.T32UN... |
Derive metadata from HDF file¶
The HDF files come with rich metadata that can be read from the files with gdalinfo. The HLS v1.4 UserGuide lists the metadata fields with a short description in Section 6.6.
The full outcome of gdalinfo for the sample scene looks as follows:
[3]:
%%bash
gdalinfo ./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf
Driver: HDF4/Hierarchical Data Format Release 4
Files: ./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf
Size is 512, 512
Metadata:
ACCODE=LaSRCL8V3.5.5; LaSRCL8V3.5.5
arop_ave_xshift(meters)=-10.5, -17.0999999999767
arop_ave_yshift(meters)=-10.5, -10.5
arop_ncp=156, 364
arop_rmse(meters)=4.32, 3.91
arop_s2_refimg=/nobackupp6/jju/S2REFIMG/32UNU_2016273A.S2A_OPER_MSI_L1C_TL_SGS__20160929T154211_A006640_T32UNU_B08.jp2
cloud_coverage=24
DATA_TYPE=L1TP; L1TP
HLS_PROCESSING_TIME=2018-05-22T01:14:35Z
HORIZONTAL_CS_NAME=UTM, WGS84, UTM ZONE 32; UTM, WGS84, UTM ZONE 32
L1_PROCESSING_TIME=2018-04-16T22:40:45Z; 2018-04-16T22:42:02Z
LANDSAT_PRODUCT_ID=LC08_L1TP_193026_20180402_20180416_01_T1; LC08_L1TP_193027_20180402_20180416_01_T1
LANDSAT_SCENE_ID=LC81930262018092LGN00; LC81930272018092LGN00
MEAN_SUN_AZIMUTH_ANGLE=154.21429006, 152.99684032
MEAN_SUN_ZENITH_ANGLE=46.61961515, 45.48706472
NBAR_Solar_Zenith=50.3345670872353
NCOLS=3660
NROWS=3660
SENSING_TIME=2018-04-02T10:03:11.3268150Z; 2018-04-02T10:03:35.2051460Z
SENSOR=OLI_TIRS; OLI_TIRS
SENTINEL2_TILEID=32UNU
spatial_coverage=29
SPATIAL_RESOLUTION=30
TIRS_SSM_MODEL=FINAL; FINAL
TIRS_SSM_POSITION_STATUS=ESTIMATED; ESTIMATED
ULX=499980
ULY=5400000
USGS_SOFTWARE=LPGS_13.0.0
Subdatasets:
SUBDATASET_1_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band01
SUBDATASET_1_DESC=[3660x3660] band01 Grid (16-bit integer)
SUBDATASET_2_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band02
SUBDATASET_2_DESC=[3660x3660] band02 Grid (16-bit integer)
SUBDATASET_3_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band03
SUBDATASET_3_DESC=[3660x3660] band03 Grid (16-bit integer)
SUBDATASET_4_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band04
SUBDATASET_4_DESC=[3660x3660] band04 Grid (16-bit integer)
SUBDATASET_5_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band05
SUBDATASET_5_DESC=[3660x3660] band05 Grid (16-bit integer)
SUBDATASET_6_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band06
SUBDATASET_6_DESC=[3660x3660] band06 Grid (16-bit integer)
SUBDATASET_7_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band07
SUBDATASET_7_DESC=[3660x3660] band07 Grid (16-bit integer)
SUBDATASET_8_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band09
SUBDATASET_8_DESC=[3660x3660] band09 Grid (16-bit integer)
SUBDATASET_9_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band10
SUBDATASET_9_DESC=[3660x3660] band10 Grid (16-bit integer)
SUBDATASET_10_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:band11
SUBDATASET_10_DESC=[3660x3660] band11 Grid (16-bit integer)
SUBDATASET_11_NAME=HDF4_EOS:EOS_GRID:"./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf":Grid:QA
SUBDATASET_11_DESC=[3660x3660] QA Grid (8-bit unsigned integer)
Corner Coordinates:
Upper Left ( 0.0, 0.0)
Lower Left ( 0.0, 512.0)
Upper Right ( 512.0, 0.0)
Lower Right ( 512.0, 512.0)
Center ( 256.0, 256.0)
The values for the fields separated from its values with a ‘=’ derived given a HDF file.
[4]:
print(df_downloaded["path"][0])
nasa_hls.get_metadata_from_hdf(df_downloaded["path"][0],
fields=["cloud_cover", "spatial_coverage"])
./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf
[4]:
{'cloud_cover': 24.0, 'spatial_coverage': 29.0}
Note that you will still get a dictionary back even if a specified field cannot be found. However you should get a warning for each field that could not be found.
[5]:
nasa_hls.get_metadata_from_hdf(df_downloaded["path"][0],
fields=["cloud_cover", "ZZZZZ", "DUBIDUBIDUUU"])
/home/ben/Devel/Repos_Py/nasa_hls/nasa_hls/utils.py:210: UserWarning: Could not find metadata for field 'ZZZZZ'.
warnings.warn(f"Could not find metadata for field '{field}'.")
/home/ben/Devel/Repos_Py/nasa_hls/nasa_hls/utils.py:210: UserWarning: Could not find metadata for field 'DUBIDUBIDUUU'.
warnings.warn(f"Could not find metadata for field '{field}'.")
[5]:
{'cloud_cover': 24.0}
Here we derive the cloud cover and spatial coverage for all the downloaded scenes.
[6]:
df_downloaded["cloud_cover"] = np.NaN
df_downloaded["spatial_coverage"] = np.NaN
for i in df_downloaded.index:
_ = nasa_hls.get_metadata_from_hdf(df_downloaded.loc[i, "path"],
fields=["cloud_cover",
"spatial_coverage"])
df_downloaded.loc[i, "cloud_cover"] = _["cloud_cover"]
df_downloaded.loc[i, "spatial_coverage"] = _["spatial_coverage"]
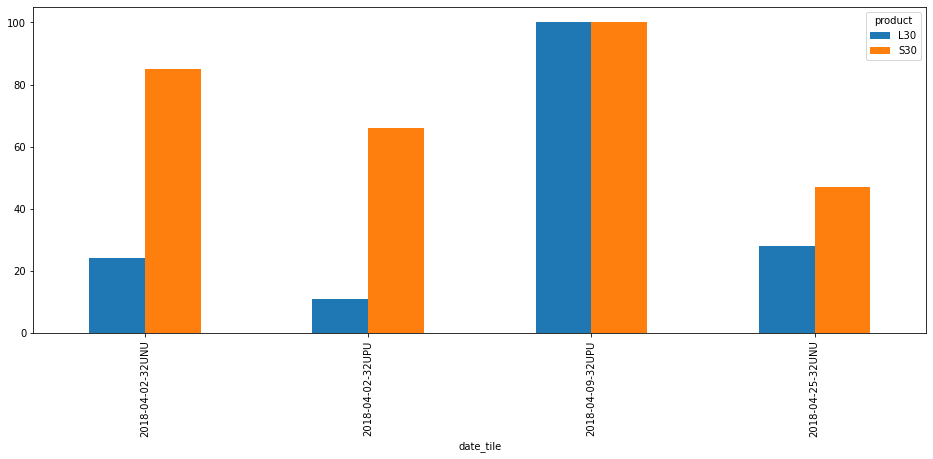
df_downloaded[["cloud_cover", "spatial_coverage"]]
[6]:
| cloud_cover | spatial_coverage | |
|---|---|---|
| id | ||
| HLS.L30.T32UNU.2018092.v1.4 | 24.0 | 29.0 |
| HLS.S30.T32UNU.2018092.v1.4 | 85.0 | 100.0 |
| HLS.L30.T32UPU.2018092.v1.4 | 11.0 | 100.0 |
| HLS.S30.T32UPU.2018092.v1.4 | 66.0 | 100.0 |
| HLS.L30.T32UPU.2018099.v1.4 | 100.0 | 50.0 |
| HLS.S30.T32UPU.2018099.v1.4 | 100.0 | 72.0 |
| HLS.L30.T32UNU.2018115.v1.4 | 28.0 | 100.0 |
| HLS.S30.T32UNU.2018115.v1.4 | 47.0 | 58.0 |
Or as a plot:
[7]:
df_downloaded["product_date"] = df_downloaded["product"] + "-" + df_downloaded["date"]
df_downloaded["date_tile"] = df_downloaded["date"] + "-" + df_downloaded["tile"]
#df_downloaded.pivot(index="product_date", columns="tile", values="cloud_cover").plot.bar(figsize=(16, 6))
df_downloaded.pivot(index="date_tile",
columns="product",
values="cloud_cover").plot.bar(figsize=(16, 6))
[7]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f5f705a8b80>
Convert HDF to single-layer GeoTIFFs¶
Note: Starting from nasa_hls version 0.1.1 you can also convert the data to Cloud Optimized GeoTIFFs (COG). This also requires GDAL >= 3.1.
To create COGs use teh option gdal_translate_options="-of COG" in convert_hdf2tiffs.
Single scene¶
[8]:
# L30
nasa_hls.convert_hdf2tiffs(Path("./xxx_uncontrolled_hls/downloads/HLS.L30.T32UNU.2018092.v1.4.hdf"),
geotiff_dir,
bands=None, # use this to convert only a subset of the bands
max_cloud_coverage=100, # use this to only convert scenes that have lower than specified cloud cover
)
[8]:
PosixPath('xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UNU.2018092.v1.4')
[9]:
# S30
nasa_hls.convert_hdf2tiffs(Path("./xxx_uncontrolled_hls/downloads/HLS.S30.T32UNU.2018092.v1.4.hdf"),
geotiff_dir,
bands=None,
max_cloud_coverage=100,
)
[9]:
PosixPath('xxx_uncontrolled_hls/geotiffs/HLS.S30.T32UNU.2018092.v1.4')
As a result you get all the (desired) bands in a subfolder of the destination directory. Note the differences between the L30 and S30 files.
[10]:
%%bash
tree xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UNU.2018092.v1.4
tree xxx_uncontrolled_hls/geotiffs/HLS.S30.T32UNU.2018092.v1.4
xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UNU.2018092.v1.4
├── HLS.L30.T32UNU.2018092.v1.4__Blue.tif
├── HLS.L30.T32UNU.2018092.v1.4__Cirrus.tif
├── HLS.L30.T32UNU.2018092.v1.4__Coastal_Aerosol.tif
├── HLS.L30.T32UNU.2018092.v1.4__Green.tif
├── HLS.L30.T32UNU.2018092.v1.4__NIR.tif
├── HLS.L30.T32UNU.2018092.v1.4__QA.tif
├── HLS.L30.T32UNU.2018092.v1.4__Red.tif
├── HLS.L30.T32UNU.2018092.v1.4__SWIR1.tif
├── HLS.L30.T32UNU.2018092.v1.4__SWIR2.tif
├── HLS.L30.T32UNU.2018092.v1.4__TIRS1.tif
└── HLS.L30.T32UNU.2018092.v1.4__TIRS2.tif
0 directories, 11 files
xxx_uncontrolled_hls/geotiffs/HLS.S30.T32UNU.2018092.v1.4
├── HLS.S30.T32UNU.2018092.v1.4__Blue.tif
├── HLS.S30.T32UNU.2018092.v1.4__Cirrus.tif
├── HLS.S30.T32UNU.2018092.v1.4__Coastal_Aerosol.tif
├── HLS.S30.T32UNU.2018092.v1.4__Green.tif
├── HLS.S30.T32UNU.2018092.v1.4__NIR_Broad.tif
├── HLS.S30.T32UNU.2018092.v1.4__NIR_Narrow.tif
├── HLS.S30.T32UNU.2018092.v1.4__QA.tif
├── HLS.S30.T32UNU.2018092.v1.4__Red_Edge1.tif
├── HLS.S30.T32UNU.2018092.v1.4__Red_Edge2.tif
├── HLS.S30.T32UNU.2018092.v1.4__Red_Edge3.tif
├── HLS.S30.T32UNU.2018092.v1.4__Red.tif
├── HLS.S30.T32UNU.2018092.v1.4__SWIR1.tif
├── HLS.S30.T32UNU.2018092.v1.4__SWIR2.tif
└── HLS.S30.T32UNU.2018092.v1.4__Water_Vapor.tif
0 directories, 14 files
Batch of scene¶
[11]:
nasa_hls.convert_hdf2tiffs_batch(df_downloaded[df_downloaded["date"]=="2018-04-02"]["path"].values,
geotiff_dir)
100%|██████████| 4/4 [00:29<00:00, 7.28s/it]
[11]:
[PosixPath('xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UNU.2018092.v1.4'),
PosixPath('xxx_uncontrolled_hls/geotiffs/HLS.S30.T32UNU.2018092.v1.4'),
PosixPath('xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UPU.2018092.v1.4'),
PosixPath('xxx_uncontrolled_hls/geotiffs/HLS.S30.T32UPU.2018092.v1.4')]
Create a clear-sky mask from the QA layer¶
The HLS v1.4 UserGuide helps to understanding and use the QA layer.
We can read in Appendix A. How to decode the bit-packed QA:
Quality Assessment (QA) encoded at the bit level provides concise presentation but is less convenient for users new to this format.
This is really true especially if compare the difficulty in handling the QA layer to handling the fmask or sen2cor SCL layer layer. Because of that and since the masking of invalid pixels / selection of clear sky pixels is so important we will go in a bit more detail here.
The rest of this section shows how to create the default mask and custom masks from the bit encoded AQ layer.
Sample QA layer¶
For illustration purposes we use the QA layer of the HLS.L30.T32UNU.2018092.v1.4 scene.
From the metadata we got the following information about the HDF file:
[12]:
df_downloaded.loc["HLS.L30.T32UNU.2018092.v1.4", ["cloud_cover", "spatial_coverage"]]
[12]:
cloud_cover 24
spatial_coverage 29
Name: HLS.L30.T32UNU.2018092.v1.4, dtype: object
Understand the QA layer¶
The metadata also gives us a QA description. We also find that information in the HLS v1.4 UserGuide:
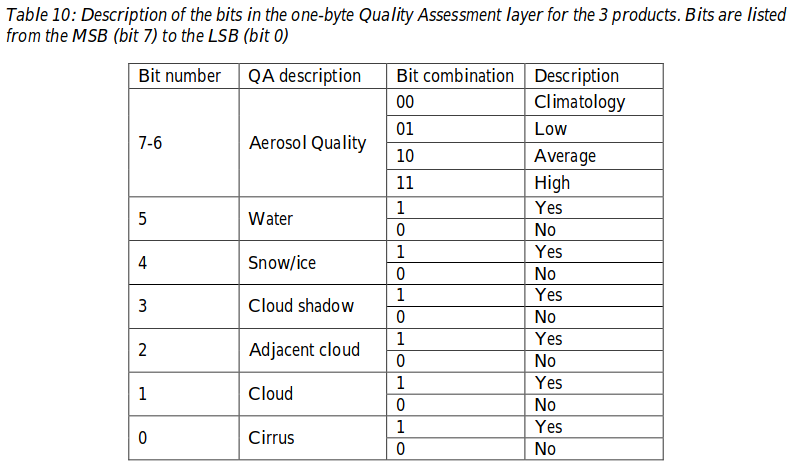
Appendix A of the HLS v1.4 UserGuide shows how to decode the QA bits with simple integer arithmetic. Here we show a way how to do this in Python.
Lets start with the example from the Appendix A:
Suppose we get a decimal QA value 100, which translates into binary 01100100, indicating that the aerosol level is low (bits 6-7), it is water (bit 5), and adjacent to cloud (bit 2).
Derive clear-sky mask from the QA layer¶
The guide gives some advice on how to use the QA layer:
Users are advised to mask out Cirrus, Cloud and Adjacent cloud pixels.
We want to find out here, which rules have been used to calculate the cloud cover we find in in the metadata. There is no explicit information about the used rules.
Therefore we first create a dataframe where the information of the table above is expanded for easier accessability. The dataframe can be created with get_qa_look_up_table.
It contains the QA value (i.e. the integer in the raster), the binary string and a boolean column which indicates if for a given QA value a given QA case (i.e. a row in the table above) is met.
[13]:
lut_qa = nasa_hls.get_qa_look_up_table()
display(lut_qa.head())
display(lut_qa.tail())
| qa_value | binary_string | a_clima | a_low | a_avg | a_high | water | snow | cloud_shadow | adj_cloud | cloud | cirrus | no_water | no_snow | no_cloud_shadow | no_adj_cloud | no_cloud | no_cirrus | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 00000000 | True | False | False | False | False | False | False | False | False | False | True | True | True | True | True | True |
| 1 | 1 | 00000001 | True | False | False | False | False | False | False | False | False | True | True | True | True | True | True | False |
| 2 | 2 | 00000010 | True | False | False | False | False | False | False | False | True | False | True | True | True | True | False | True |
| 3 | 3 | 00000011 | True | False | False | False | False | False | False | False | True | True | True | True | True | True | False | False |
| 4 | 4 | 00000100 | True | False | False | False | False | False | False | True | False | False | True | True | True | False | True | True |
| qa_value | binary_string | a_clima | a_low | a_avg | a_high | water | snow | cloud_shadow | adj_cloud | cloud | cirrus | no_water | no_snow | no_cloud_shadow | no_adj_cloud | no_cloud | no_cirrus | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 251 | 251 | 11111011 | False | False | False | True | True | True | True | False | True | True | False | False | False | True | False | False |
| 252 | 252 | 11111100 | False | False | False | True | True | True | True | True | False | False | False | False | False | False | True | True |
| 253 | 253 | 11111101 | False | False | False | True | True | True | True | True | False | True | False | False | False | False | True | False |
| 254 | 254 | 11111110 | False | False | False | True | True | True | True | True | True | False | False | False | False | False | False | True |
| 255 | 255 | 11111111 | False | False | False | True | True | True | True | True | True | True | False | False | False | False | False | False |
Based on that dataframe it is easy to get the QA integer values for creating a specific mask from it.
Find masking rules resulting in metadata cloud cover¶
As an example let us see if we can calculate the cloud cover from the QA layer by finding the right QA attributes to be masked.
Let’s get the QA values of the sample scene.
[14]:
qa_path = './xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UNU.2018092.v1.4/HLS.L30.T32UNU.2018092.v1.4__QA.tif'
with rasterio.open(qa_path) as qa:
meta = qa.meta
qa_array = qa.read()
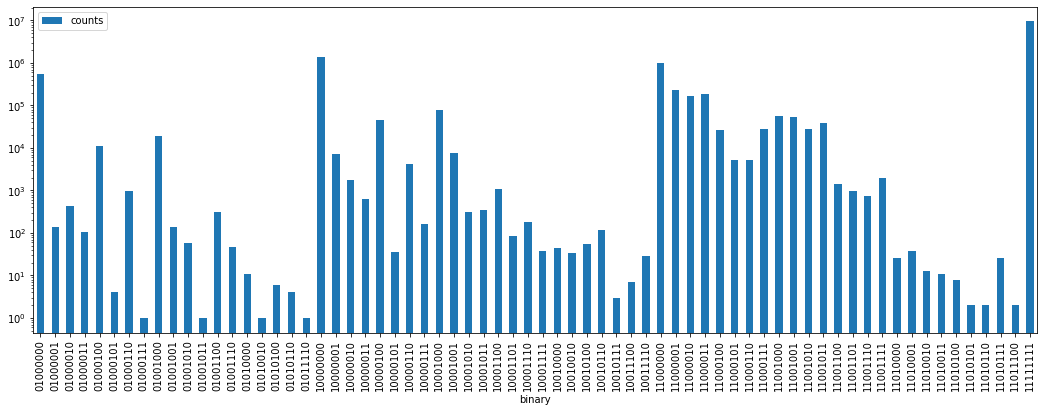
Let’s first visualize the distribution of the values:
[15]:
qa_counts = pd.DataFrame(np.unique(qa_array), columns=["integer"])
qa_counts["binary"] = qa_counts.integer.apply(lambda i: "{0:08b}".format(i))
qa_counts["counts"] = qa_counts.integer.apply(lambda i: (qa_array == i).sum())
n_pix = qa_counts["counts"].sum()
n_obs = qa_counts[qa_counts["integer"] != 255].counts.sum()
print("number of observations (not 255) :", n_obs)
qa_counts.plot.bar(x="binary", y="counts", logy=True, figsize=(18, 6))
number of observations (not 255) : 3930335
[15]:
<matplotlib.axes._subplots.AxesSubplot at 0x7f5f7029ee50>
Let’s also calculate the percentage of pixels with respect to all pixels and with respect to the observed pixels (i.e. not 255):
[16]:
qa_counts["percent"] = ((qa_counts["counts"] / n_pix) * 100).round(2)
qa_counts["percent_obs"] = ((qa_counts["counts"] / n_obs) * 100).round(2)
qa_counts.loc[qa_counts["integer"] == 255, "percent_obs"] = -0.01
qa_counts.tail()
qa_counts_top_22 = qa_counts.sort_values("percent_obs", ascending=False).head(22)
qa_counts_top_22
[16]:
| integer | binary | counts | percent | percent_obs | |
|---|---|---|---|---|---|
| 19 | 128 | 10000000 | 1389745 | 10.37 | 35.36 |
| 42 | 192 | 11000000 | 991982 | 7.41 | 25.24 |
| 0 | 64 | 01000000 | 535713 | 4.00 | 13.63 |
| 43 | 193 | 11000001 | 228818 | 1.71 | 5.82 |
| 45 | 195 | 11000011 | 185779 | 1.39 | 4.73 |
| 44 | 194 | 11000010 | 170090 | 1.27 | 4.33 |
| 27 | 136 | 10001000 | 79370 | 0.59 | 2.02 |
| 50 | 200 | 11001000 | 57441 | 0.43 | 1.46 |
| 51 | 201 | 11001001 | 53104 | 0.40 | 1.35 |
| 23 | 132 | 10000100 | 44661 | 0.33 | 1.14 |
| 53 | 203 | 11001011 | 38433 | 0.29 | 0.98 |
| 49 | 199 | 11000111 | 28472 | 0.21 | 0.72 |
| 52 | 202 | 11001010 | 28155 | 0.21 | 0.72 |
| 46 | 196 | 11000100 | 26763 | 0.20 | 0.68 |
| 8 | 72 | 01001000 | 18869 | 0.14 | 0.48 |
| 4 | 68 | 01000100 | 10947 | 0.08 | 0.28 |
| 28 | 137 | 10001001 | 7772 | 0.06 | 0.20 |
| 20 | 129 | 10000001 | 7303 | 0.05 | 0.19 |
| 47 | 197 | 11000101 | 5277 | 0.04 | 0.13 |
| 48 | 198 | 11000110 | 5087 | 0.04 | 0.13 |
| 25 | 134 | 10000110 | 4114 | 0.03 | 0.10 |
| 57 | 207 | 11001111 | 1991 | 0.01 | 0.05 |
[17]:
df_downloaded.loc["HLS.L30.T32UNU.2018092.v1.4", ["cloud_cover", "spatial_coverage"]]
[17]:
cloud_cover 24
spatial_coverage 29
Name: HLS.L30.T32UNU.2018092.v1.4, dtype: object
Spatial coverage
[18]:
print("spatial coverage from QA: ", 100 - qa_counts.loc[qa_counts["integer"] == 255, "percent"].values[0])
spatial coverage from QA: 29.340000000000003
Cloud cover
I could not find in the docs how the AQ layer was used to calculate the cloud cover.
With the percentages in qa_counts and the lut_qa dataframe it is now easy to see how different masking decisions influence the percentage of masked pixels for a given QA layer.
The following shows my guesses to find a match for a cloud cover of 24 %.
[19]:
valid = lut_qa.loc[
~((lut_qa.cirrus) | (lut_qa.cloud) | (lut_qa.adj_cloud)), "qa_value"
].values
percent_valid = qa_counts.loc[qa_counts.integer.isin(valid), "percent_obs"].sum()
100 - percent_valid
[19]:
21.810000000000002
[20]:
valid = lut_qa.loc[
~((lut_qa.cloud) | (lut_qa.adj_cloud) | (lut_qa.cloud_shadow)), "qa_value"
].values # => 19.8
percent_valid = qa_counts.loc[qa_counts.integer.isin(valid), "percent_obs"].sum()
100 - percent_valid
[20]:
19.75999999999999
[21]:
valid = lut_qa.loc[
~((lut_qa.cirrus) | (lut_qa.cloud) | (lut_qa.adj_cloud) | (lut_qa.cloud_shadow)), "qa_value"
].values # => 25.8
percent_valid = qa_counts.loc[qa_counts.integer.isin(valid), "percent_obs"].sum()
100 - percent_valid
[21]:
25.769999999999996
[22]:
valid = lut_qa.loc[
~((lut_qa.cirrus) | (lut_qa.cloud) | (lut_qa.cloud_shadow)), "qa_value"
].values # => 23.7
percent_valid = qa_counts.loc[qa_counts.integer.isin(valid), "percent_obs"].sum()
100 - percent_valid
[22]:
23.67
The winner is: cirrus, cloud and cloud shadow, which somehow makes sense.
That means that we consider the following QA values as clear sky or valid pixels:
[23]:
valid
[23]:
array([ 0, 4, 16, 20, 32, 36, 48, 52, 64, 68, 80, 84, 96,
100, 112, 116, 128, 132, 144, 148, 160, 164, 176, 180, 192, 196,
208, 212, 224, 228, 240, 244])
Convert the QA layer into a mask¶
Given we know the QA values that we want to consider as valid or invalid pixels we can derive a mask array or GeoTIFF from a GeoTIFF QA layer.
Here we use the function hls_qa_layer_to_mask to quickly test if the masking with the valid values give us the same cloud cover as the mateadata.
Note that you have the following additional options:
keep_255(bool): IfTruethis keeps the value 255 for the non sensed pixels.mask_path(str): If a path is given the function will create a raster of the mask there and not output an array.overwrite(bool): Defines if a file existing atmask_pathshould be overwritten.
[24]:
for i, row in df_downloaded.iterrows():
sceneid = Path(row.path).stem
qa_path = f'xxx_uncontrolled_hls/geotiffs/{sceneid}/{sceneid}__QA.tif'
if not Path(qa_path).exists():
continue
clear_arr = nasa_hls.hls_qa_layer_to_mask(qa_path,
qa_valid=valid,
keep_255=True,
mask_path=None,
overwrite=False)
n_pix = clear_arr.size
n_obs = (clear_arr != 255).sum()
n_cloudy = (clear_arr == 0).sum()
n_clear = (clear_arr == 1).sum()
cloud_cover = n_cloudy / (n_clear + n_cloudy)
print("*" * 90)
print(qa_path)
print(f"Number of pixels : {n_pix}")
print(f"Number of obs. : {n_obs}")
print(f"Number of clear obs. : {n_clear}")
print(f"Number of cloudy obs. : {n_cloudy}")
print(f"Cloud cover - metadata : {row.cloud_cover}")
print(f"Cloud cover - calculated : {round(cloud_cover * 100, 1) }")
print(f"Spatial coverage - metadata : {row.spatial_coverage}")
print(f"Spatial coverage - calculated: {round((n_obs / n_pix) * 100, 1) }")
******************************************************************************************
xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UNU.2018092.v1.4/HLS.L30.T32UNU.2018092.v1.4__QA.tif
Number of pixels : 13395600
Number of obs. : 3930335
Number of clear obs. : 2999963
Number of cloudy obs. : 930372
Cloud cover - metadata : 24.0
Cloud cover - calculated : 23.7
Spatial coverage - metadata : 29.0
Spatial coverage - calculated: 29.3
******************************************************************************************
xxx_uncontrolled_hls/geotiffs/HLS.S30.T32UNU.2018092.v1.4/HLS.S30.T32UNU.2018092.v1.4__QA.tif
Number of pixels : 13395600
Number of obs. : 13272703
Number of clear obs. : 1976262
Number of cloudy obs. : 11296441
Cloud cover - metadata : 85.0
Cloud cover - calculated : 85.1
Spatial coverage - metadata : 100.0
Spatial coverage - calculated: 99.1
******************************************************************************************
xxx_uncontrolled_hls/geotiffs/HLS.L30.T32UPU.2018092.v1.4/HLS.L30.T32UPU.2018092.v1.4__QA.tif
Number of pixels : 13395600
Number of obs. : 13395365
Number of clear obs. : 11980540
Number of cloudy obs. : 1414825
Cloud cover - metadata : 11.0
Cloud cover - calculated : 10.6
Spatial coverage - metadata : 100.0
Spatial coverage - calculated: 100.0
******************************************************************************************
xxx_uncontrolled_hls/geotiffs/HLS.S30.T32UPU.2018092.v1.4/HLS.S30.T32UPU.2018092.v1.4__QA.tif
Number of pixels : 13395600
Number of obs. : 12915488
Number of clear obs. : 4495212
Number of cloudy obs. : 8420276
Cloud cover - metadata : 66.0
Cloud cover - calculated : 65.2
Spatial coverage - metadata : 100.0
Spatial coverage - calculated: 96.4
!!! NOTE AND WARNING !!!
There are some small irregularities between the metadata and the calculated values that should be investigated further.
Feel free to do so… (;
Visualize the mask¶
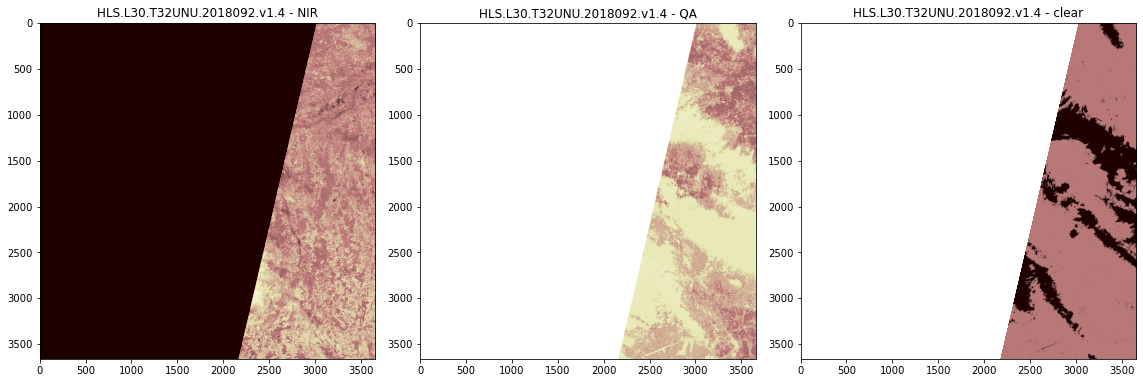
[25]:
sceneid = "HLS.L30.T32UNU.2018092.v1.4"
src_nir = rasterio.open(f'xxx_uncontrolled_hls/geotiffs/{sceneid}/{sceneid}__NIR.tif')
src_qa = rasterio.open(f'xxx_uncontrolled_hls/geotiffs/{sceneid}/{sceneid}__QA.tif')
qa_path = f'xxx_uncontrolled_hls/geotiffs/{sceneid}/{sceneid}__QA.tif'
clear_arr = nasa_hls.hls_qa_layer_to_mask(qa_path,
qa_valid=valid,
keep_255=True,
mask_path=None,
overwrite=False)
fig, axes = plt.subplots(1, 3, figsize=(16, 8))
axes[0].imshow(src_nir.read(1), vmin=0, vmax=6000, cmap='pink')
axes[0].set_title(sceneid + " - " + "NIR")
axes[1].imshow(src_qa.read(1), vmin=0, vmax=255, cmap='pink')
axes[1].set_title(sceneid + " - " + "QA")
axes[2].imshow(clear_arr, vmin=0, vmax=3, cmap='pink')
axes[2].set_title(sceneid + " - " + "clear")
fig.tight_layout()