Comparing USGS ShakeMap versions
The M 7.1 - 72 km ENE of Namie, Japan earthquake.
Overview
Recently, I came across the question if and how it is possible to download superseded versions of ShakeMaps distributed by the USGS Earthquake Hazards program. This post comprises what I encountered on my first walk through the rich resources offered by the USGS.
In a nutshell, we will
- use the libcomcat Python library to
- query the history of product submissions (e.g. ShakeMaps) related to an event
- download different versions of a product (here the raster.zip)
- reprocess rasters to have common extend and pixel alignment
- visualize how the Modified Mercalli Intensity (MMI) changes over different product versions for a given event.
We will walk through these steps using the M 7.1 - 72 km ENE of Namie, Japan event.
Data and Tools
The USGS Earthquake Hazards Program > Data and Tools webpage provides an overview of available data and different ways to access them. The Real-time Notifications, Feeds, and Web Services webpage lists different ways to access real-time notifications as well as some links for developers.
We can find a link to the web service API Documentation - Earthquake Catalog there. But we can also click our way through the Developers Corner to the Python libcomcat project that provides a Python equivalent to the ANSS ComCat search API, i.e. the API Documentation - Earthquake Catalog.
In this post, we will use the libcomcat Python API. The Python libcomcat README.md contains links to the API Documentation, the Command Line Interface Documentation, and several great Jupyter notebooks that make it easy to get started with the API. Most of the code snippets in this post are taken from there.
Libraries
from datetime import timezone
from libcomcat.dataframes import get_history_data_frame, split_history_frame
from libcomcat.search import get_event_by_id
import matplotlib.pyplot as plt
import numpy as np
from pathlib import Path
import pandas as pd
import rasterio
from rasterio.warp import reproject, Resampling
import zipfile
pd.set_option('display.max_columns', None)
Event Details
Assume we already know the event ID of an earthquake of interest.
Then we can get a DetailEvent instance for it.
quake_id = 'us6000dher'
event = get_event_by_id(quake_id, includesuperseded=True)
event
us6000dher 2021-02-13 14:07:50.397000 (37.745,141.749) 49.9 km M7.1
With this object, we can derive the history of the event’s products in a pandas data frame with the get_history_data_frame function.
We need to create the object with includesuperseded=True to get all the versions of the event’s product (History Dataframe).
df_products, event = get_history_data_frame(event)
display(df_products.head(3))
df_products['Product'].value_counts()
| Update Time | Product | Authoritative Event ID | Code | Associated | Product Source | Product Version | Elapsed (min) | URL | Comment | Description | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 313 | 2021-02-13 14:19:04.892 | origin | us6000dher | pt21044000 | True | pt | 1 | 11.2 | https://earthquake.usgs.gov/archive/product/or... | Magnitude# 7.1|Time# 2021-02-13 14:07:00 |Time... | |
| 311 | 2021-02-13 14:20:10.768 | origin | us6000dher | at00qoh0l1 | True | at | 1 | 12.3 | https://earthquake.usgs.gov/archive/product/or... | Magnitude# 7.1|Time# 2021-02-13 14:07:52 |Time... | |
| 312 | 2021-02-13 14:20:10.774 | origin | us6000dher | at00qoh0l1 | True | at | 2 | 12.3 | https://earthquake.usgs.gov/archive/product/or... | Magnitude# 7.1|Time# 2021-02-13 14:07:52 |Time... |
dyfi 285
moment-tensor 10
ground-failure 8
losspager 7
shakemap 7
origin 7
phase-data 4
finite-fault 1
Name: Product, dtype: int64
The value counts on the Products column show that there are seven ShakeMap versions.
We can subset the dataset by hand or a dedicated helper function.
The helper function will subset the data, reset the row index and change the columns of the data frame.
df_shakemap = split_history_frame(df_products, product='shakemap')
df_shakemap
| Update Time | Product | Authoritative Event ID | Code | Associated | Product Source | Product Version | Elapsed (min) | URL | Comment | MaxMMI | Instrumented | DYFI | Fault | GMPE | Mag | Depth | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2021-02-13 14:28:21.006 | shakemap | us6000dher | us6000dher | True | us | 1 | 20.5 | https://earthquake.usgs.gov/archive/product/sh... | 5.2 | 0.0 | 0.0 | NaN | [AtkinsonBoore2003SInter],[ZhaoEtAl2016SInter]... | 7.0 | 54.0 | |
| 1 | 2021-02-13 15:10:42.406 | shakemap | us6000dher | us6000dher | True | us | 2 | 62.9 | https://earthquake.usgs.gov/archive/product/sh... | 6.4 | 4.0 | 23.0 | NaN | [AtkinsonBoore2003SInter],[ZhaoEtAl2016SInter]... | 7.0 | 54.0 | |
| 2 | 2021-02-13 16:11:24.776 | shakemap | us6000dher | us6000dher | True | us | 3 | 123.6 | https://earthquake.usgs.gov/archive/product/sh... | 6.5 | 4.0 | 36.0 | NaN | [AtkinsonBoore2003SInter],[ZhaoEtAl2016SInter]... | 7.1 | 50.6 | |
| 3 | 2021-02-13 22:11:55.217 | shakemap | us6000dher | us6000dher | True | us | 4 | 484.1 | https://earthquake.usgs.gov/archive/product/sh... | 7.0 | 4.0 | 43.0 | NaN | [AbrahamsonEtAl2014],[BooreEtAl2014],[Campbell... | 7.1 | 35.0 | |
| 4 | 2021-02-14 00:17:56.940 | shakemap | us6000dher | us6000dher | True | us | 5 | 610.1 | https://earthquake.usgs.gov/archive/product/sh... | 7.6 | 589.0 | 43.0 | NaN | [AbrahamsonEtAl2014],[BooreEtAl2014],[Campbell... | 7.1 | 35.0 | |
| 5 | 2021-02-14 14:12:31.505 | shakemap | us6000dher | us6000dher | True | us | 6 | 1444.7 | https://earthquake.usgs.gov/archive/product/sh... | 8.0 | 586.0 | 49.0 | NaN | [AbrahamsonEtAl2014],[BooreEtAl2014],[Campbell... | 7.1 | 35.0 | |
| 6 | 2021-02-14 22:08:57.989 | shakemap | us6000dher | us6000dher | True | us | 7 | 1921.1 | https://earthquake.usgs.gov/archive/product/sh... | 8.0 | 586.0 | 50.0 | NaN | [AbrahamsonEtAl2014],[BooreEtAl2014],[Campbell... | 7.1 | 49.9 |
ShakeMap Product
Inspect contents
The ShakeMap product comprises several contents or files.
The Product class instance has methods that make it easy to list and download specific contents or files.
Let us look at the Product instance of the preferred version of the ShakeMap product.
products = event.getProducts('shakemap', version='preferred')
print(products) # list of products but we only expect one due to version='preferred'
product = products[0]
print(f'Contents: {product.contents}')
[Product shakemap from us updated 2021-02-13 15:10:42.406000 containing 58 content files.]
Contents: ['contents.xml', 'download/attenuation_curves.json', 'download/cont_mi.json', 'download/cont_mmi.json', 'download/cont_pga.json', 'download/cont_pgv.json', 'download/cont_psa0p3.json', 'download/cont_psa1p0.json', 'download/cont_psa3p0.json', 'download/coverage_mmi_high_res.covjson', 'download/coverage_mmi_low_res.covjson', 'download/coverage_mmi_medium_res.covjson', 'download/coverage_pga_high_res.covjson', 'download/coverage_pga_low_res.covjson', 'download/coverage_pga_medium_res.covjson', 'download/coverage_pgv_high_res.covjson', 'download/coverage_pgv_low_res.covjson', 'download/coverage_pgv_medium_res.covjson', 'download/coverage_psa0p3_high_res.covjson', 'download/coverage_psa0p3_low_res.covjson', 'download/coverage_psa0p3_medium_res.covjson', 'download/coverage_psa1p0_high_res.covjson', 'download/coverage_psa1p0_low_res.covjson', 'download/coverage_psa1p0_medium_res.covjson', 'download/coverage_psa3p0_high_res.covjson', 'download/coverage_psa3p0_low_res.covjson', 'download/coverage_psa3p0_medium_res.covjson', 'download/grid.xml', 'download/info.json', 'download/intensity.jpg', 'download/intensity.pdf', 'download/intensity_overlay.png', 'download/intensity_overlay.pngw', 'download/mmi_legend.png', 'download/mmi_regr.png', 'download/pga.jpg', 'download/pga.pdf', 'download/pga_regr.png', 'download/pgv.jpg', 'download/pgv.pdf', 'download/pgv_regr.png', 'download/pin-thumbnail.png', 'download/psa0p3.jpg', 'download/psa0p3.pdf', 'download/psa0p3_regr.png', 'download/psa1p0.jpg', 'download/psa1p0.pdf', 'download/psa1p0_regr.png', 'download/psa3p0.jpg', 'download/psa3p0.pdf', 'download/psa3p0_regr.png', 'download/raster.zip', 'download/rupture.json', 'download/shake_result.hdf', 'download/shakemap.kmz', 'download/shape.zip', 'download/stationlist.json', 'download/uncertainty.xml']
This list shows 58 files associated with a specific version of the ShakeMap product. These can also be found and downloaded under the respective event page.
Download content
Now, we will download the raster.zip. Since we will download different versions, we build a destination directory containing the product version. Then we extract the zip file.
path_zip = Path(f'data/{quake_id}_{product.source}_{product.version:02d}_raster.zip')
path_zip.parent.mkdir(parents=True, exist_ok=True)
print('Zip file local:', path_zip)
print('Source URL:')
print(product.getContent('raster.zip', path_zip))
with zipfile.ZipFile(path_zip, 'r') as zip_ref:
zip_ref.extractall(path_zip.parent / path_zip.stem)
Zip file local: data/us6000dher_us_02_raster.zip
Source URL:
https://earthquake.usgs.gov/archive/product/shakemap/us6000dher/us/1613229042406/download/raster.zip
Now we can do the same thing for all versions.
We could also do this via the CLI with
getproduct shakemap raster.zip -i us6000dher --get-version all
def download_and_extract_raster(product, quake_id, basedir, overwrite=False):
path_zip = Path(f'{str(basedir)}/{quake_id}_{product.source}_{product.version:02d}_raster.zip')
path_zip.parent.mkdir(parents=True, exist_ok=True)
print('Zip file local:', path_zip)
if not path_zip.exists() or overwrite:
print('Source URL:',
product.getContent('raster.zip', path_zip))
with zipfile.ZipFile(path_zip, 'r') as zip_ref:
zip_ref.extractall(path_zip.parent / path_zip.stem)
for product in event.getProducts('shakemap', version='all'):
print(f'****** {product.version} ******')
download_and_extract_raster(product, quake_id, basedir='data', overwrite=False)
****** 1 ******
Zip file local: data/us6000dher_us_01_raster.zip
Source URL: https://earthquake.usgs.gov/archive/product/shakemap/us6000dher/us/1613226501006/download/raster.zip
****** 2 ******
Zip file local: data/us6000dher_us_02_raster.zip
****** 3 ******
Zip file local: data/us6000dher_us_03_raster.zip
Source URL: https://earthquake.usgs.gov/archive/product/shakemap/us6000dher/us/1613232684776/download/raster.zip
****** 4 ******
Zip file local: data/us6000dher_us_04_raster.zip
Source URL: https://earthquake.usgs.gov/archive/product/shakemap/us6000dher/us/1613254315217/download/raster.zip
****** 5 ******
Zip file local: data/us6000dher_us_05_raster.zip
Source URL: https://earthquake.usgs.gov/archive/product/shakemap/us6000dher/us/1613261876940/download/raster.zip
****** 6 ******
Zip file local: data/us6000dher_us_06_raster.zip
Source URL: https://earthquake.usgs.gov/archive/product/shakemap/us6000dher/us/1613311951505/download/raster.zip
****** 7 ******
Zip file local: data/us6000dher_us_07_raster.zip
Source URL: https://earthquake.usgs.gov/archive/product/shakemap/us6000dher/us/1613340537989/download/raster.zip
With the update_time we can generate the number in the URL leading to the respective versions.
For example, in the case of the last product we downloaded:
str(product.update_time.replace(tzinfo=timezone.utc).timestamp()).replace('.','')
'1613340537989'
Reproject and stack rasters
For intended visualization, we need the different rasters to fit. To see if this is the case, we can investigate the metadata of different raster files.
First, let us get a list of files we want to compare and see if we need to reproject them.
paths_mmi = list(Path('data').rglob('mmi_mean.flt'))
paths_mmi.sort()
paths_mmi
[PosixPath('data/us6000dher_us_01_raster/mmi_mean.flt'),
PosixPath('data/us6000dher_us_02_raster/mmi_mean.flt'),
PosixPath('data/us6000dher_us_03_raster/mmi_mean.flt'),
PosixPath('data/us6000dher_us_04_raster/mmi_mean.flt'),
PosixPath('data/us6000dher_us_05_raster/mmi_mean.flt'),
PosixPath('data/us6000dher_us_06_raster/mmi_mean.flt'),
PosixPath('data/us6000dher_us_07_raster/mmi_mean.flt')]
rasterio.open(paths_mmi[0]).meta
{'driver': 'EHdr',
'dtype': 'float32',
'nodata': 999.0,
'width': 498,
'height': 394,
'count': 1,
'crs': None,
'transform': Affine(0.016666666666666666, 0.0, 138.025,
0.0, -0.016666666666666666, 40.891666666666666)}
rasterio.open(paths_mmi[-1]).meta
{'driver': 'EHdr',
'dtype': 'float32',
'nodata': 999.0,
'width': 511,
'height': 427,
'count': 1,
'crs': None,
'transform': Affine(0.016666666666666666, 0.0, 135.49166666666667,
0.0, -0.016666666666666666, 41.208333333333336)}
The rasters of the different versions do not match, as we can see from the width, height, and transform values. Therefore reprojection is necessary. We will use the last version to define our template or destination array and reproject all the other versions to match it.
The implementation here follows rasterio’s Reprojection documentation. We directly reproject raster values loaded into a numpy array object. The documentation also describes and references ways to reproject raster files.
Let us first configure the destination array:
mmi_template = rasterio.open(paths_mmi[-1])
dst_shape = mmi_template.shape
dst_transform = mmi_template.transform
dst_crs = {'init': 'EPSG:4326'}
destination = np.zeros(mmi_template.shape, mmi_template.meta['dtype'])
With that, we can reproject the data of the other versions to match it and collect all versions in a three-dimensional array.
mmi_mean_version_stack = []
for i, path_mmi_i in enumerate(paths_mmi[:-1]):
mmi_i = rasterio.open(path_mmi_i)
src_shape = mmi_i.shape
src_transform = mmi_i.transform
src_crs = {'init': 'EPSG:4326'}
source = mmi_i.read()
mmi_mean_version_stack.append(
reproject(
source,
destination,
src_transform=src_transform,
src_crs=src_crs,
dst_transform=dst_transform,
dst_crs=dst_crs,
resampling=Resampling.nearest)[0] # returns tuple (arrray(...), Affine(...))
)
mmi_mean_version_stack.append(mmi_template.read()[0, :, :])
mmi_mean_version_stack = np.stack(mmi_mean_version_stack, axis=2)
mmi_mean_version_stack.shape
(427, 511, 7)
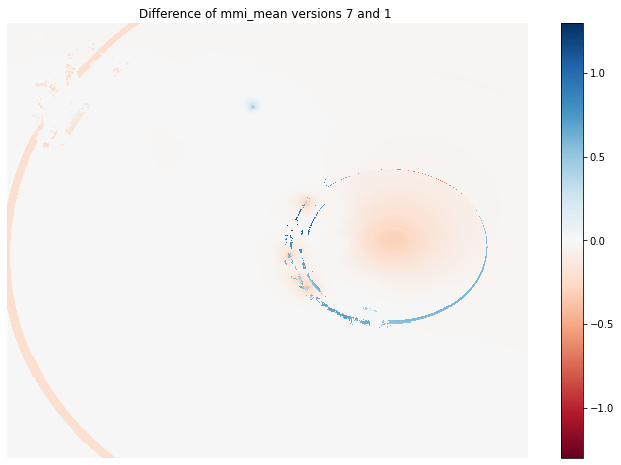
Visual comparison of two ShakeMap versions
Finally, we have the different raster versions in a format that allows us to easily analyze how ShakeMap versions differ.
For example, the following plot shows the difference between the first and the last versions of mmi_mean.
fig, axes = plt.subplots(1,1, figsize=(12, 8))
diff_v7v1 = mmi_mean_version_stack[:, :, -1] - mmi_mean_version_stack[:, :, 0]
img1 = axes.imshow(
diff_v7v1,
cmap ='RdBu',
vmin=-1.3,
vmax=1.3,
interpolation ='nearest',
origin ='lower')
axes.set_title('Difference of mmi_mean versions 7 and 1 ')
axes.axis('off')
cbar = fig.colorbar(img1)